Cytoscape Alternatives. Cytoscape is described as 'open source software platform for visualizing complex networks and integrating these with any type of attribute data' and is an app in the Social & Communications category. There are more than 10 alternatives to Cytoscape for a variety of platforms, including Windows, Linux, Mac, the Web and iPad.
i am trying to implement cytoscape in my lab and have installed it on the centralized server which we access via ssh from own terminals (with x11 forwarding).
- Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression.
- #vue-cytoscape # What is this This is a vue wrapper for cytoscape.js. # Why We all love vue for creating user interfaces and cytoscape.js as a graph visualization library. Wouldn't be nice if we can create a reusablevue component that exposes cytoscape.js api?
- Cytoscape is an open source software project for integrating biomolecular interaction networks with high-throughput expression data and other molecular states into a unified conceptual framework. Although applicable to any system of molecular components and interactions, Cytoscape is most powerful w.
Cytoscape Api
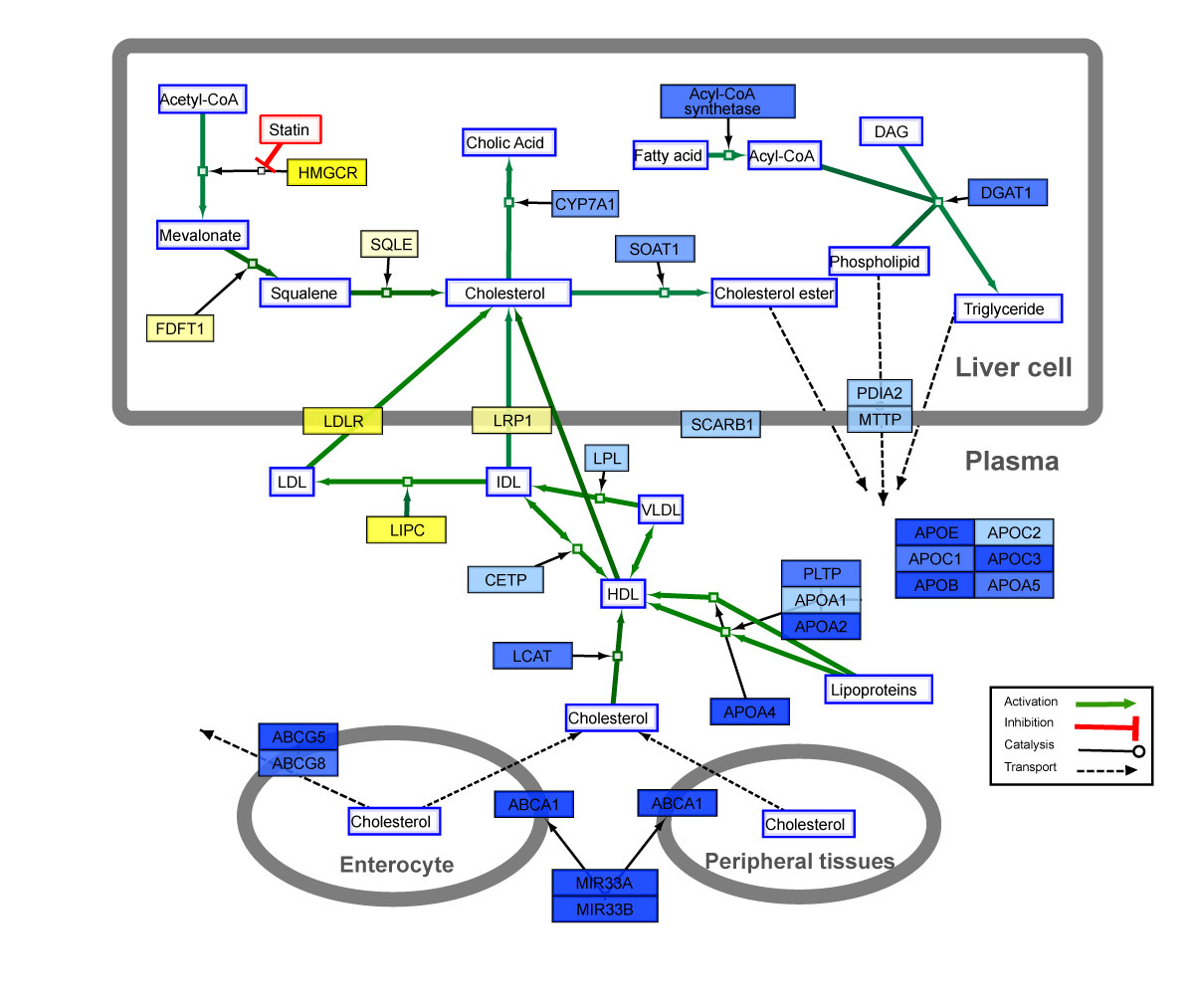
currently cytoscape 3.8.2 is installed (as root) in /opt/ and executing it as non-root users works. however, if a second user launches it while an instance is already running, it throws error:
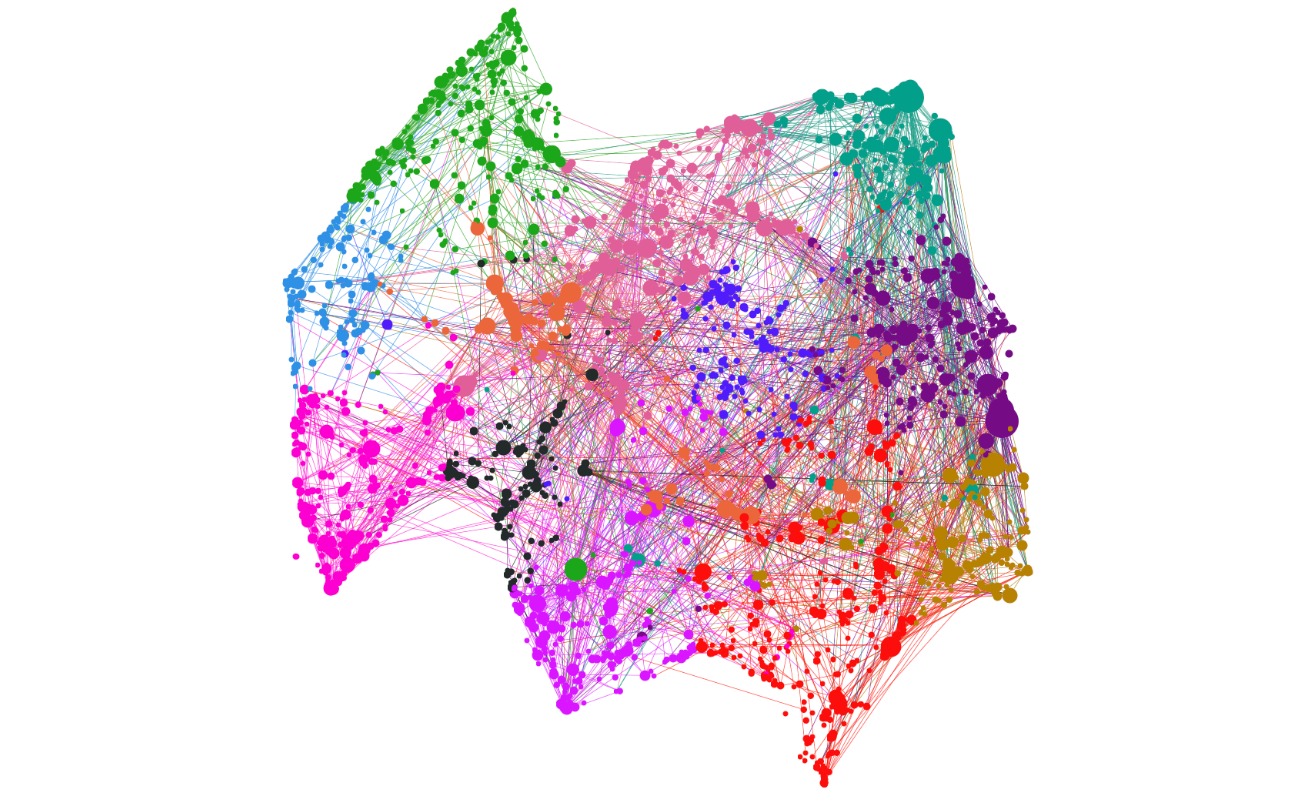
Cytoscape Software
karaf: There is a Root instance already running with name Cytoscape 3.8.2 and pid 121496. If you know what you are doing and want to force the run anyway, export CHECK_ROOT_INSTANCE_RUNNING=false and re run the command.
is there a way to have a centralised installation of cytoscape and have multiple users access it from their own terminals?
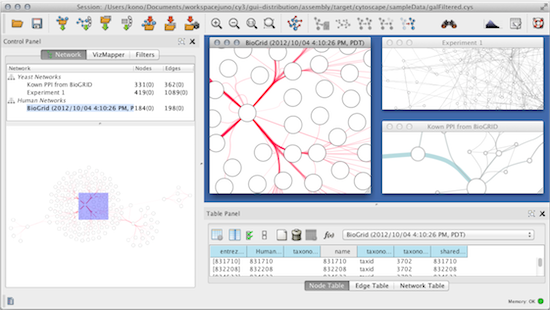
or is it safe to do:
Cytoscape Web
CHECK_ROOT_INSTANCE_RUNNING=false
